Grid Processing Tools
Grid Processing Tools
PCP-Combine
PCP-Combine
Functionality
The PCP-Combine tool is used (if needed) to sum, add, or subtract accumulated precipitation from one or more gridded data files into a single NetCDF file containing the desired accumulation period. Its NetCDF output may be used as input to the MET statistics tools. It may be run to sum the same accumulation interval across multiple files, to add data from one or more files, or to subtract data from exactly two files. The sum, add, and subtract options may be run on any gridded data file supported by MET. The input files being combined must have already been placed on the grid on which the user would like to verify. The copygb and wgrib2 utilities are recommended for re-gridding GRIB1 and GRIB2 files, respectively. In addition, the PCP-Combine tool will only sum model files with the same initialization time unless it is told to ignore the initialization time.
PCP-Combine Usage
View the usage statement for PCP-Combine by simply typing the following:
Use the -sum, -add, -subtract, or -derive command line option to indicate the operation to be performed. Each operation has its own set of required arguments.
By default, PCP-Combine searches for records containing GRIB code 61 (accumulated precipitation). Use the -config config_str option to override this default and process other fields.
Run
Run
Since PCP-Combine performs a simple operation and reformatting step, no configuration file is needed.
Run PCP-Combine on the command line using the following two commands:
-sum 20050807_000000 3 20050808_000000 12 \
$MET_TUTORIAL_DATA/output/pcp_combine/sample_fcst_24L_2005080800V_12A.nc \
-pcpdir $MET_TUTORIAL_DATA/input/sample_fcst/2005080700
pcp_combine \
-sum 00000000_000000 1 20050808_000000 12 \
$MET_TUTORIAL_DATA/output/pcp_combine/sample_obs_2005080800V_12A.nc \
-pcpdir $MET_TUTORIAL_DATA/input/sample_obs/ST2ml
In the first command, PCP-Combine summed up 4 3-hourly accumulation forecast files into a single 12-hour accumulation forecast. In the second command, PCP-Combine summed up 12 1-hourly accumulation observation files into a single 12-hour accumulation observation. PCP-Combine should perform these tasks very quickly. In the second command, we told PCP-Combine to ignore the initialization time by setting its YYYYMMDD_HHMMSS to all zeros (0000000_000000).
Output
Output
When PCP-Combine is finished, you may view the output NetCDF files it wrote using the ncdump and ncview utilities (if available on your machine). Run the following commands to view contents of the NetCDF files:
ncview $MET_TUTORIAL_DATA/output/pcp_combine/sample_obs_2005080800V_12A.nc &
ncdump -h $MET_TUTORIAL_DATA/output/pcp_combine/sample_fcst_24L_2005080800V_12A.nc
ncdump -h $MET_TUTORIAL_DATA/output/pcp_combine/sample_obs_2005080800V_12A.nc
The ncview windows display plots of the precipitation data in these files. The output of ncdump indicates that the gridded fields are named APCP_12, the GRIB code abbreviation and accumulation interval for precipitation. We'll use these NetCDF output files from PCP-Combine as input for the Grid-Stat tool.
Gen-Vx-Mask
Gen-Vx-Mask
Gen-Vx-Mask Tool: General
Gen-Vx-Mask Functionality
The Gen-Vx-Mask tool is an optional utility that may be run one or more times to define a verification masking region for a domain. The MET tools provide a variety of ways to define spatial verification masking regions over which to accumulate statistics. One such way is to define a polyline consisting of Lat/Lon points which define the region. The Lat/Lon polyline files may be used directly in the other MET tools, however, doing so, for a relatively complex polyline with a few hundred points is inefficient. The Gen-Vx-Mask tool may be used instead to apply a polyline to a domain once and define a bitmap for the mask. Using the output of Gen-Vx-Mask to define verification masking regions in other MET tools is much more efficient.
The Gen-Vx-Mask tool replaces the Gen-Poly-Mask and Gen-Circle-Mask tools from earlier versions and adds support for several ways of defining masking regions, described below. This tool may be run iteratively to combine multiple masks together.
Gen-Vx-Mask Usage
View the usage statement for Gen-Vx-Mask by simply typing the following:
At a minimum, the input_file, the mask_file, and the NetCDF out_file must be passed in on the command line. The input_fileis a gridded data file which defines the domain for which the mask should be defined. The NetCDF out_file is the output NetCDF mask file to be written. The mask_file defines the masking information:
- For "poly", "box", "circle", and "track" masking, specify an ASCII Lat/Lon file.
- For "grid" and "data" masking, specify a gridded data file.
- For "solar_alt" and "solar_azi" masking, specify a gridded data file or a timestring in YYYYMMDD[_HH[MMSS]] format.
- For "lat" and "lon" masking, no "mask_file" needed, simply repeat the path for "input_file".
The Gen-Vx-Mask tool supports the following settings for the -type command line option:
- For -type poly (default), the mask_file is a Lat/Lon polyline file which defines the area to be included in the mask.
- For -type box, the mask_file is a Lat/Lon polyline file which defines individual locations of interest. The -height and -width options define the number of grid points surrounding that location to be included in the mask.
- For -type circle, the mask_file is a Lat/Lon polyline file which defines individual locations of interest. For each grid point, the minimum distance in kilometers to the set of masking points is computed. If the -thresh option is provided, those distances will be thresholded to define a mask. Otherwise, the actual distance values will be written.
- The -type track option is very similar to circle. However, the Lat/Lon points define a connected track rather than individual locations. For each grid point, the minimum distance in kilometers to that track is computed. If the -thresh option is provided, those distances will be thresholded to define a mask. Otherwise, the actual distance values will be written.
- For -type grid, the mask_file is a gridded data file whose grid defines the desired masking region. Any grid points falling inside the masking grid are included in the mask.
- For -type data, the mask_file is a gridded data file which should be on the same grid as in_file. The -mask_field option specifies the field of data which should be extracted from the mask_file. If the -thresh option is provided, those data values will be thresholded to define a mask. Otherwise, the actual data values will be written.
- The -type solar_alt specifies the position of the solar altitude.
- The -type solar_azi specifies the azimuth of the sun.
- The -type lat computes the latitude value at each grid point. This logic only requires the definition of the grid, specified by the input_file. Technically, the mask_file is not needed, but a value must be specified for the command line to parse correctly. Users are advised to simply repeat the input_file setting twice.
- The -type lon computes the longitude value at each grid point. This logic only requires the definition of the grid, specified by the input_file. Technically, the mask_file is not needed, but a value must be specified for the command line to parse correctly. Users are advised to simply repeat the input_file setting twice.
- The -type shape option and value specifies shapefile masking using a closed polygon taken from an ESRI shapefile to define the masking region. Gen-Vx-Mask reads the shapefile with the ".shp" suffix and extracts the latitude and longitude of the verticies. The other types of shapefiles (index file, suffix ".shx" and dBASE file, suffix ".dbf") are not currently used. The shapefile must consist of closed polygons rather than polylines, points, or any of the other data types that shapefiles support. Shapefiles usually contain more than one polygon and the -shapeno n command line option enables the user to select one polygone from the shapefile. The integer ntells which shape number to use from the shapefile. Note that this value is zero-based, so that the first polygone in the shapefile is polygon number 0, the second is 1, etc. For the user's convenience, some utilities that perform human-readable screen dumps of shapefile contents are provided. The gis_dump_shp, gis_dump_shx, and gis_dump_dbf tools enable the user to examine the contents of the shapefiles.
The Gen-Vx-Mask tool supports several other useful options:
- The input_file defines the domain of interest, and by default, each grid point is assigned an initial value of 0. The -input_field option may be used to specify a field of data whose values override the default. The input value at each grid point will be written to the output unless its value is updated during the current masking process.
- The -mask_field option is for "data" masking, define the field from "mask_file" to be used.
- The -complement option may be used to apply the opposite (i.e. reverse video) of the current mask. For example, for -type polymasking, all grid points outside the polyline will be selected instead of all points inside.
- The -union, -intersection, and -symdiff options define the logic for combining values from the -input_field with the current mask values.
- By default, 1 is the value written to the output for a grid point inside the mask. The -value option overrides that default value.
- The -name option overrides the default variable name written to the NetCDF output file.
- The -height and -width options are for "box" masking, specify these dimensions in grid units.
- The -thresh defines the threshold to be applied:
- For "circle" and "track" masking, threshold the distance (km).
- For "data" masking, threshold the values of "mask_field".
- For "solar_alt" and "solar_azi" masking, threshold the computed solar values.
- For "lat" and "lon" masking, threshold the latitude and longitude values.
The variety of options for the Gen-Vx-Mask tool and the ability to run it iteratively enable the user great flexibility in defining the spatial areas for computing statistics.
Run
Run
Since Gen-Vx-Mask performs a relatively simple masking operation, no configuration file is needed.
Run Gen-Vx-Mask to apply polyline masking using the following command:
-type poly \
$MET_TUTORIAL_DATA/input/sample_obs/ST2ml/ST2ml2005080700.Grb_G212 \
$MET_BASE/poly/CONUS.poly \
$MET_TUTORIAL_DATA/output/gen_vx_mask/CONUS_G212_poly.nc
In this command, Gen-Vx-Mask reads a gridded data file on the NCEP Grid 212 domain and a Lat/Lon polyline file defining a rough outline of the contiguous United States. Gen-Vx-Mask extracts the domain information from the data file, applies the CONUS polyline, and writes out a NetCDF mask file.
Run Gen-Vx-Mask to apply circle masking using the following command:
-type circle -thresh '<=200' \
$MET_TUTORIAL_DATA/input/sample_fcst/2005080700/wrfprs_ruc13_24.tm00_G212 \
$MET_BASE/poly/MLB_Stadiums.txt \
$MET_TUTORIAL_DATA/output/gen_vx_mask/circle_mask.nc
In this command, Gen-Vx-Mask reads a gridded data file that's on the NCEP Grid 212 domain, computes the minimum distance to each Lat/Lon location defined in the MLB_Stadiums.txt file, and applies a 200 km distance threshold. It creates a NetCDF file named circle_mask.nc containing a bitmap for the domain with a value of 1 for all grid points inside the circles and a value of 0 for all grid points outside.
Output
Output
When Gen-Vx-Mask is finished, you may view the output NetCDF file it wrote using the ncdump and ncview utilities (if available on your machine). Run the following commands to view the contents of the NetCDF files:
ncdump -h $MET_TUTORIAL_DATA/output/gen_vx_mask/CONUS_G212_poly.nc
ncview $MET_TUTORIAL_DATA/output/gen_vx_mask/circle_mask.nc &
ncdump -h $MET_TUTORIAL_DATA/output/gen_vx_mask/circle_mask.nc
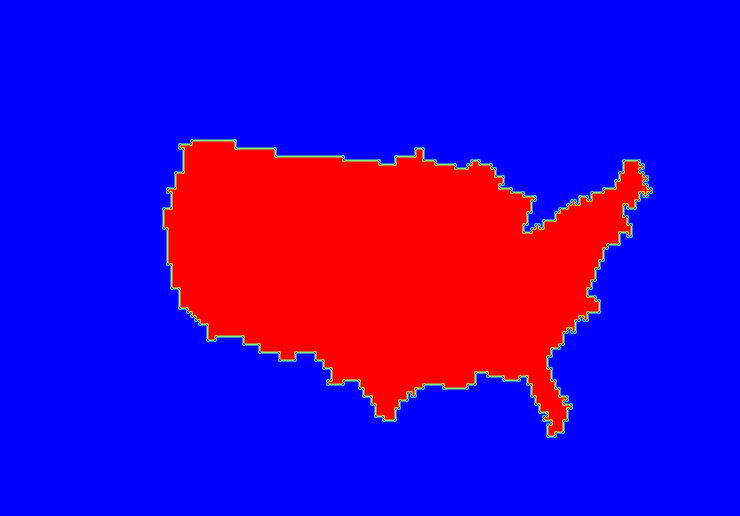
The output of ncdump indicates that the NetCDF file contains a variable named CONUS, which is the name defined for this polyline in the input Lat/Lon file. The ncview window should display a plot similar to Figure 2 below. We'll use this NetCDF output file from Gen-Vx-Mask to define verification masking regions for the other MET tools.
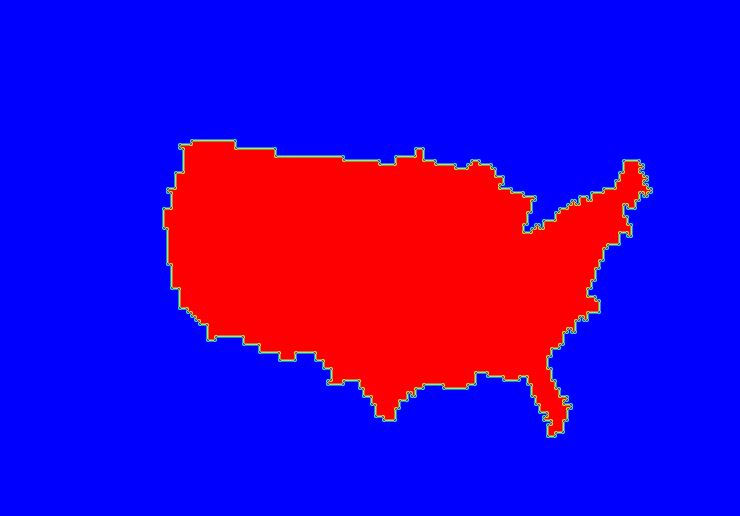
The output of ncdump indicates that the NetCDF file contains a variable named MLB_Stadiums, which is the name defined for this masking region in the input Lat/Lon file. The ncview window should display a plot similar to Figure 3 below.
Additional Examples
The following examples illustrate the definition of more complex masks. After running each example, use ncview to display the results.
- Change the threshold for circle masking:
-type circle -thresh '>=400' \
$MET_TUTORIAL_DATA/input/sample_fcst/2005080700/wrfprs_ruc13_24.tm00_G212 \
$MET_BASE/poly/MLB_Stadiums.txt \
$MET_TUTORIAL_DATA/output/gen_vx_mask/circle_ge400_mask.nc
-
Apply data masking where surface pressure is below 850mb and intersect with the CONUS mask:
-type data -intersection \
$MET_TUTORIAL_DATA/output/gen_vx_mask/CONUS_G212_poly.nc \
-input_field 'name="CONUS"; level="(*,*)";' \
$MET_TUTORIAL_DATA/input/sample_fcst/2005080700/wrfprs_ruc13_12.tm00_G212 \
-mask_field 'name="PRES"; level="Z0";' -thresh 'le85000' \
$MET_TUTORIAL_DATA/output/gen_vx_mask/sfcpres_850_and_conus_mask.nc
- Apply box masking and complement :
-type box -height 5 -width 10 -complement \
$MET_TUTORIAL_DATA/input/sample_fcst/2005080700/wrfprs_ruc13_24.tm00_G212 \
$MET_BASE/poly/MLB_Stadiums.txt \
$MET_TUTORIAL_DATA/output/gen_vx_mask/box_complement_mask.nc
- Compute track distances with no threshold:
-type track \
$MET_TUTORIAL_DATA/input/sample_fcst/2005080700/wrfprs_ruc13_24.tm00_G212 \
$MET_BASE/poly/MLB_Stadiums.txt \
$MET_TUTORIAL_DATA/output/gen_vx_mask/track_distance.nc